update
This commit is contained in:
148
pages/CSE559A/CSE559A_L10.md
Normal file
148
pages/CSE559A/CSE559A_L10.md
Normal file
@@ -0,0 +1,148 @@
|
||||
# CSE559A Lecture 10
|
||||
|
||||
## Convolutional Neural Networks
|
||||
|
||||
### Convolutional Layer
|
||||
|
||||
Output feature map resolution depends on padding and stride
|
||||
|
||||
Padding: add zeros around the input image
|
||||
|
||||
Stride: the step of the convolution
|
||||
|
||||
Example:
|
||||
|
||||
1. Convolutional layer for 5x5 image with 3x3 kernel, padding 1, stride 1 (no skipping pixels)
|
||||
- Input: 5x5 image
|
||||
- Output: 3x3 feature map, (5-3+2*1)/1+1=5
|
||||
2. Convolutional layer for 5x5 image with 3x3 kernel, padding 1, stride 2 (skipping pixels)
|
||||
- Input: 5x5 image
|
||||
- Output: 2x2 feature map, (5-3+2*1)/2+1=2
|
||||
|
||||
_Learned weights can be thought of as local templates_
|
||||
|
||||
```python
|
||||
import torch
|
||||
import torch.nn as nn
|
||||
|
||||
# suppose input image is HxWx3 (assume RGB image)
|
||||
|
||||
conv_layer = nn.Conv2d(in_channels=3, # input channel, input is HxWx3
|
||||
out_channels=64, # output channel (number of filters), output is HxWx64
|
||||
kernel_size=3, # kernel size
|
||||
padding=1, # padding, this ensures that the output feature map has the same resolution as the input image, H_out=H_in, W_out=W_in
|
||||
stride=1) # stride
|
||||
```
|
||||
|
||||
Usually followed by a ReLU activation function
|
||||
|
||||
```python
|
||||
conv_layer = nn.Conv2d(in_channels=3, out_channels=64, kernel_size=3, padding=1, stride=1)
|
||||
relu = nn.ReLU()
|
||||
```
|
||||
|
||||
Suppose input image is $H\times W\times K$, the output feature map is $H\times W\times L$ with kernel size $F\times F$, this takes $F^2\times K\times L\times H\times W$ parameters
|
||||
|
||||
Each operation $D\times (K^2C)$ matrix with $(K^2C)\times N$ matrix, assume $D$ filters and $C$ output channels.
|
||||
|
||||
### Variants 1x1 convolutions, depthwise convolutions
|
||||
|
||||
#### 1x1 convolutions
|
||||
|
||||
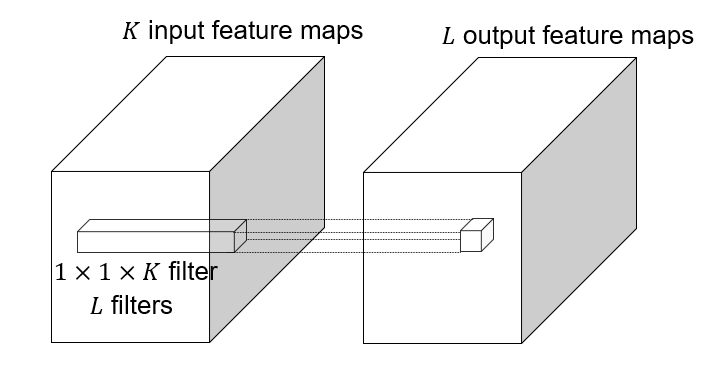
|
||||
|
||||
1x1 convolution: $F=1$, this layer do convolution in the pixel level, it is **pixel-wise** convolution for the feature.
|
||||
|
||||
Used to save computation, reduce the number of parameters.
|
||||
|
||||
Example: 3x3 conv layer with 256 channels at input and output.
|
||||
|
||||
Option 1: naive way:
|
||||
|
||||
```python
|
||||
conv_layer = nn.Conv2d(in_channels=256, out_channels=256, kernel_size=3, padding=1, stride=1)
|
||||
```
|
||||
|
||||
This takes $256\times 3 \times 3\times 256=524,288$ parameters.
|
||||
|
||||
Option 2: 1x1 convolution:
|
||||
|
||||
```python
|
||||
conv_layer = nn.Conv2d(in_channels=256, out_channels=64, kernel_size=1, padding=0, stride=1)
|
||||
conv_layer = nn.Conv2d(in_channels=64, out_channels=64, kernel_size=3, padding=1, stride=1)
|
||||
conv_layer = nn.Conv2d(in_channels=64, out_channels=256, kernel_size=1, padding=0, stride=1)
|
||||
```
|
||||
|
||||
This takes $256\times 1\times 1\times 64 + 64\times 3\times 3\times 64 + 64\times 1\times 1\times 256 = 16,384 + 36,864 + 16,384 = 69,632$ parameters.
|
||||
|
||||
This lose some information, but save a lot of parameters.
|
||||
|
||||
#### Depthwise convolutions
|
||||
|
||||
Depthwise convolution: $K\to K$ feature map, save computation, reduce the number of parameters.
|
||||
|
||||
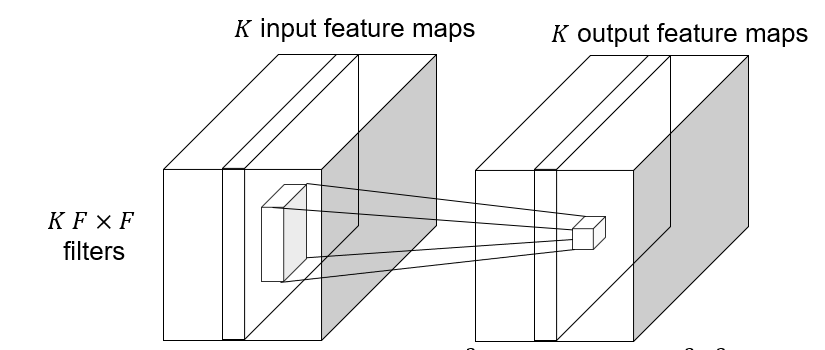
|
||||
|
||||
#### Grouped convolutions
|
||||
|
||||
Self defined convolution on the feature map following the similar manner.
|
||||
|
||||
### Backward pass
|
||||
|
||||
Vector-matrix form:
|
||||
|
||||
$$
|
||||
\frac{\partial e}{\partial x}=\frac{\partial e}{\partial z}\frac{\partial z}{\partial x}
|
||||
$$
|
||||
|
||||
Suppose the kernel is 3x3, the feature map is $\ldots, x_{i-1}, x_i, x_{i+1}, \ldots$, and $\ldots, z_{i-1}, z_i, z_{i+1}, \ldots$ is the output feature map, then:
|
||||
|
||||
The convolution operation can be written as:
|
||||
|
||||
$$
|
||||
z_i = w_1x_{i-1} + w_2x_i + w_3x_{i+1}
|
||||
$$
|
||||
|
||||
The gradient of the kernel is:
|
||||
|
||||
$$
|
||||
\frac{\partial e}{\partial x_i} = \sum_{j=-1}^{1}\frac{\partial e}{\partial z_i}\frac{\partial z_i}{\partial x_i} = \sum_{j=-1}^{1}\frac{\partial e}{\partial z_i}w_j
|
||||
$$
|
||||
|
||||
### Max-pooling
|
||||
|
||||
Get max value in the local region.
|
||||
|
||||
#### Receptive field
|
||||
|
||||
The receptive field of a unit is the region of the input feature map whose values contribute to the response of that unit (either in the previous layer or in the initial image)
|
||||
|
||||
## Architecture of CNNs
|
||||
|
||||
### AlexNet (2012-2013)
|
||||
|
||||
Successor of LeNet-5, but with a few significant changes
|
||||
|
||||
- Max pooling, ReLU nonlinearity
|
||||
- Dropout regularization
|
||||
- More data and bigger model (7 hidden layers, 650K units, 60M params)
|
||||
- GPU implementation (50x speedup over CPU)
|
||||
- Trained on two GPUs for a week
|
||||
|
||||
#### Key points
|
||||
|
||||
Most floating point operations occur in the convolutional layers.
|
||||
|
||||
Most of the memory usage is in the early convolutional layers.
|
||||
|
||||
Nearly all parameters are in the fully-connected layers.
|
||||
|
||||
### VGGNet (2014)
|
||||
|
||||
### GoogLeNet (2014)
|
||||
|
||||
### ResNet (2015)
|
||||
|
||||
### Beyond ResNet (2016 and onward): Wide ResNet, ResNeXT, DenseNet
|
||||
|
||||
|
||||
@@ -12,4 +12,5 @@ export default {
|
||||
CSE559A_L7: "Computer Vision (Lecture 7)",
|
||||
CSE559A_L8: "Computer Vision (Lecture 8)",
|
||||
CSE559A_L9: "Computer Vision (Lecture 9)",
|
||||
CSE559A_L10: "Computer Vision (Lecture 10)",
|
||||
}
|
||||
|
||||
190
pages/Math416/Math416_L10.md
Normal file
190
pages/Math416/Math416_L10.md
Normal file
@@ -0,0 +1,190 @@
|
||||
# Math416 Lecture 10
|
||||
|
||||
## Fast reload on Power Series
|
||||
|
||||
Suppose $\sum_{n=0}^\infty a_n$ converges absolutely. ($\sum_{n=0}^\infty |a_n|<\infty$)
|
||||
|
||||
Then rearranging the terms of the series does not affect the sum of the series.
|
||||
|
||||
For any permutation $\sigma$ of the set of positive integers, $\sum_{n=0}^\infty a_{\sigma(n)}=\sum_{n=0}^\infty a_n$.
|
||||
|
||||
Proof:
|
||||
|
||||
Let $\epsilon>0$, then $\exists N\in\mathbb{N}$ such that $\forall n\geq N$,
|
||||
|
||||
$$
|
||||
\sum_{n=N}^\infty |a_n|<\epsilon
|
||||
$$
|
||||
|
||||
So there exists $N_0$ such that if $M\geq N_0$, then
|
||||
|
||||
$$
|
||||
\sum_{n=N_0}^M |a_n|<\epsilon
|
||||
$$
|
||||
|
||||
_for any first $M$ terms of $\sigma$, we choose $N_0$ such that all the terms (no overlapping with the first $M$ terms) on the tail is less than $\epsilon$_.
|
||||
|
||||
$$
|
||||
\sum_{n=1}^{\infty} a_n=\sum_{n=1}^{M} a_n+\sum_{n=M+1}^\infty a_n
|
||||
$$
|
||||
|
||||
Let $K>N$, $L>N_0$,
|
||||
|
||||
$$
|
||||
\left|\sum_{n=1}^{K}a_n-\sum_{n=1}^{L}a_{\sigma(n)}\right|<2\epsilon
|
||||
$$
|
||||
|
||||
EOP
|
||||
|
||||
## Chapter 4 Complex Integration
|
||||
|
||||
### Complex Integral
|
||||
|
||||
#### Definition 4.1
|
||||
|
||||
If $\phi(t)$ is a complex function defined on $[a,b]$, then the integral of $\phi(t)$ over $[a,b]$ is defined as
|
||||
|
||||
$$
|
||||
\int_a^b \phi(t) dt = \int_a^b \text{Re}\{\phi(t)\} dt + i\int_a^b \text{Im}\{\phi(t)\} dt
|
||||
$$
|
||||
|
||||
#### Theorem 4.3 (Triangle Inequality)
|
||||
|
||||
If $\phi(t)$ is a complex function defined on $[a,b]$, then
|
||||
|
||||
$$
|
||||
\left|\int_a^b \phi(t) dt\right| \leq \int_a^b |\phi(t)| dt
|
||||
$$
|
||||
|
||||
Proof:
|
||||
|
||||
Let $\lambda(t)=\frac{\left|\int_a^t \phi(t) dt\right|}{\int_a^t |\phi(t)| dt}$, then $\left|\lambda(t)\right|=1$.
|
||||
|
||||
$$
|
||||
\begin{aligned}
|
||||
\left|\int_a^b \phi(t) dt\right|&=\lambda\int_a^b \phi(t) dt\\
|
||||
&=\int_a^b \lambda(t)\phi(t) dt\\
|
||||
&=\text{Re} \{\int_a^b \lambda(t)\phi(t) dt\}\\
|
||||
&\leq\int_a^b |\lambda(t)\phi(t)| dt\\
|
||||
&=\int_a^b |\phi(t)| dt
|
||||
\end{aligned}
|
||||
$$
|
||||
|
||||
Assume $\phi$ is continuous on $[a,b]$, the equality means $\lambda(t)\phi(t)$ is real and positive everywhere on $[a,b]$, which means $\arg \phi(t)$ is constant.
|
||||
|
||||
EOP
|
||||
|
||||
#### Definition 4.4 Arc Length
|
||||
|
||||
Let $\gamma$ be a curve in the complex plane defined by $\gamma(t)=x(t)+iy(t)$, $t\in[a,b]$. The arc length of $\gamma$ is given by
|
||||
|
||||
$$
|
||||
\Gamma=\int_a^b |\gamma'(t)| dt=\int_a^b \sqrt{\left(\frac{dx}{dt}\right)^2+\left(\frac{dy}{dt}\right)^2} dt
|
||||
$$
|
||||
|
||||
N.B. If $\int_{\Gamma} f(\zeta) d\zeta$ depends on orientation of $\Gamma$, but not the parametrization.
|
||||
|
||||
We define
|
||||
|
||||
$$
|
||||
\int_{\Gamma} f(\zeta) d\zeta=\int_{\Gamma} f(\gamma(t))\gamma'(t) dt
|
||||
$$
|
||||
|
||||
Example:
|
||||
|
||||
Suppose $\Gamma$ is the circle centered at $z_0$ with radius $R$
|
||||
|
||||
$$
|
||||
\int_{\Gamma} \frac{1}{\zeta-z_0} d\zeta
|
||||
$$
|
||||
|
||||
Parameterize the unit circle:
|
||||
|
||||
$$
|
||||
\gamma(t)=z_0+Re^{it}\quad
|
||||
\gamma'(t)=iRe^{it}, t\in[0,2\pi]
|
||||
$$
|
||||
|
||||
$$
|
||||
f(\zeta)=\frac{1}{\zeta-z_0}
|
||||
$$
|
||||
|
||||
$$
|
||||
f(\gamma(t))=\frac{1}{(z_0+Re^{it})-z_0}
|
||||
$$
|
||||
|
||||
$$
|
||||
\int_{\Gamma} f(\zeta) d\zeta=\int_0^{2\pi} f(\gamma(t))\gamma'(t) dt=\int_0^{2\pi} \frac{1}{Re^{-it}}iRe^{it} dt=2\pi i
|
||||
$$
|
||||
|
||||
#### Theorem 4.11 (Uniform Convergence)
|
||||
|
||||
If $f_n(z)$ converges uniformly to $f(z)$ on $\Gamma$, assume length of $\Gamma$ is finite, then
|
||||
|
||||
$$
|
||||
\lim_{n\to\infty} \int_{\Gamma} f_n(z) dz = \int_{\Gamma} f(z) dz
|
||||
$$
|
||||
|
||||
Proof:
|
||||
|
||||
Let $\epsilon>0$, since $f_n(z)$ converges uniformly to $f(z)$ on $\Gamma$, there exists $N\in\mathbb{N}$ such that for all $n\geq N$,
|
||||
|
||||
$$
|
||||
\left|f_n(z)-f(z)\right|<\epsilon
|
||||
$$
|
||||
|
||||
$$
|
||||
\begin{aligned}
|
||||
\left|\int_{\Gamma} f_n(z) dz - \int_{\Gamma} f(z) dz\right|&=\left|\int_{\Gamma} (f_n(\gamma(t))-f(\gamma(t)))\gamma'(t) dt\right|\\
|
||||
&\leq \int_{\Gamma} |f_n(\gamma(t))-f(\gamma(t))||\gamma'(t)| dt\\
|
||||
&\leq \int_{\Gamma} \epsilon|\gamma'(t)| dt\\
|
||||
&=\epsilon\text{length}(\Gamma)
|
||||
\end{aligned}
|
||||
$$
|
||||
|
||||
EOP
|
||||
|
||||
#### Theorem 4.6 (Integral of derivative)
|
||||
|
||||
Suppose $\Gamma$ is a closed curve, $\gamma:[a,b]\to\mathbb{C}$ and $\gamma(a)=\gamma(b)$.
|
||||
|
||||
$$
|
||||
\begin{aligned}
|
||||
\int_{\Gamma} f'(z) dz &= \int_a^b f'(\gamma(t))\gamma'(t) dt\\
|
||||
&=\int_a^b \frac{d}{dt}f(\gamma(t)) dt\\
|
||||
&=f(\gamma(b))-f(\gamma(a))\\
|
||||
&=0
|
||||
\end{aligned}
|
||||
$$
|
||||
|
||||
EOP
|
||||
|
||||
Example:
|
||||
|
||||
Let $R$ be a rectangle $\{-a,a,ai+b,ai-b\}$, $\Gamma$ is the boundary of $R$ with positive orientation.
|
||||
|
||||
Let $\int_{R} e^{-\zeta^2}d\zeta$.
|
||||
|
||||
Is $e^{-\zeta^2}=\frac{d}{d\zeta}f(\zeta)$?
|
||||
|
||||
Yes, since
|
||||
|
||||
$$
|
||||
e^{\zeta^2}=1-\frac{\zeta^2}{1!}+\frac{\zeta^4}{2!}-\frac{\zeta^6}{3!}+\cdots=\frac{d}{d\zeta}\left(\frac{\zeta}{1!}-\frac{1}{3}\frac{\zeta^3}{2!}+\frac{1}{5}\frac{\zeta^5}{3!}-\cdots\right)
|
||||
$$
|
||||
|
||||
This is polynomial, therefore holomorphic.
|
||||
|
||||
So
|
||||
|
||||
$$
|
||||
\int_{R} e^{\zeta^2}d\zeta = 0
|
||||
$$
|
||||
|
||||
with some limit calculation, we can get
|
||||
|
||||
<!--TODO: Fill the parts-->
|
||||
|
||||
$$
|
||||
\int_{R} e^{-\zeta^2}d\zeta = 2\pi i
|
||||
$$
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 3
|
||||
# Math416 Lecture 3
|
||||
|
||||
## Differentiation of functions in complex variables
|
||||
|
||||
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 4
|
||||
# Math416 Lecture 4
|
||||
|
||||
## Review
|
||||
|
||||
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 5
|
||||
# Math416 Lecture 5
|
||||
|
||||
## Review
|
||||
|
||||
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 6
|
||||
# Math416 Lecture 6
|
||||
|
||||
## Review
|
||||
|
||||
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 7
|
||||
# Math416 Lecture 7
|
||||
|
||||
## Review
|
||||
|
||||
|
||||
@@ -1,4 +1,4 @@
|
||||
# Lecture 8
|
||||
# Math416 Lecture 8
|
||||
|
||||
## Review
|
||||
|
||||
|
||||
@@ -11,5 +11,6 @@ export default {
|
||||
Math416_L6: "Complex Variables (Lecture 6)",
|
||||
Math416_L7: "Complex Variables (Lecture 7)",
|
||||
Math416_L8: "Complex Variables (Lecture 8)",
|
||||
Math416_L9: "Complex Variables (Lecture 9)"
|
||||
Math416_L9: "Complex Variables (Lecture 9)",
|
||||
Math416_L10: "Complex Variables (Lecture 10)",
|
||||
}
|
||||
|
||||
@@ -2,4 +2,4 @@
|
||||
|
||||
This is a course about symmetrical group and bunch of applications in other fields of math.
|
||||
|
||||
Prof. Fere is teaching this course.
|
||||
Prof. Renado Fere is teaching this course.
|
||||
|
||||
BIN
public/CSE559A/1x1_layer.png
Normal file
BIN
public/CSE559A/1x1_layer.png
Normal file
Binary file not shown.
|
After Width: | Height: | Size: 14 KiB |
BIN
public/CSE559A/Depthwise_layer.png
Normal file
BIN
public/CSE559A/Depthwise_layer.png
Normal file
Binary file not shown.
|
After Width: | Height: | Size: 22 KiB |
Reference in New Issue
Block a user